SPOP is detected as a mutational cancer driver in Prostate Adenocarcinoma
SPOP reports in Prostate Adenocarcinoma (PRAD)
Cancer type details
| Prostate Adenocarcinoma |
|---|
|
Cohorts
13
Samples 2,819 Mutations 9,128,298 Driver genes 104 |
Gene details
| SPOP |
|---|
|
Ensembl ID
ENSG00000121067
Transcript ID ENST00000393328 Protein ID ENSP00000377001 |
|
Cohorts where is driver
12
Mutated samples 214 Mutated samples (%) 7.59 Mutations 220 |
| Mode of action Activating |
| Known driver True |
Method signals per Cohort
| Cohort | Methods | Samples | Samples (%) |
|---|
ClustL
HotMAPS
smRegions
Clustered Mutations
CBaSE dNdScv Recurrent Mutations
FML Functional Mutations
MutPanning Tri-nucleotide specific bias
combination Combination
CBaSE dNdScv Recurrent Mutations
FML Functional Mutations
MutPanning Tri-nucleotide specific bias
combination Combination
In-silico saturation mutagenesis
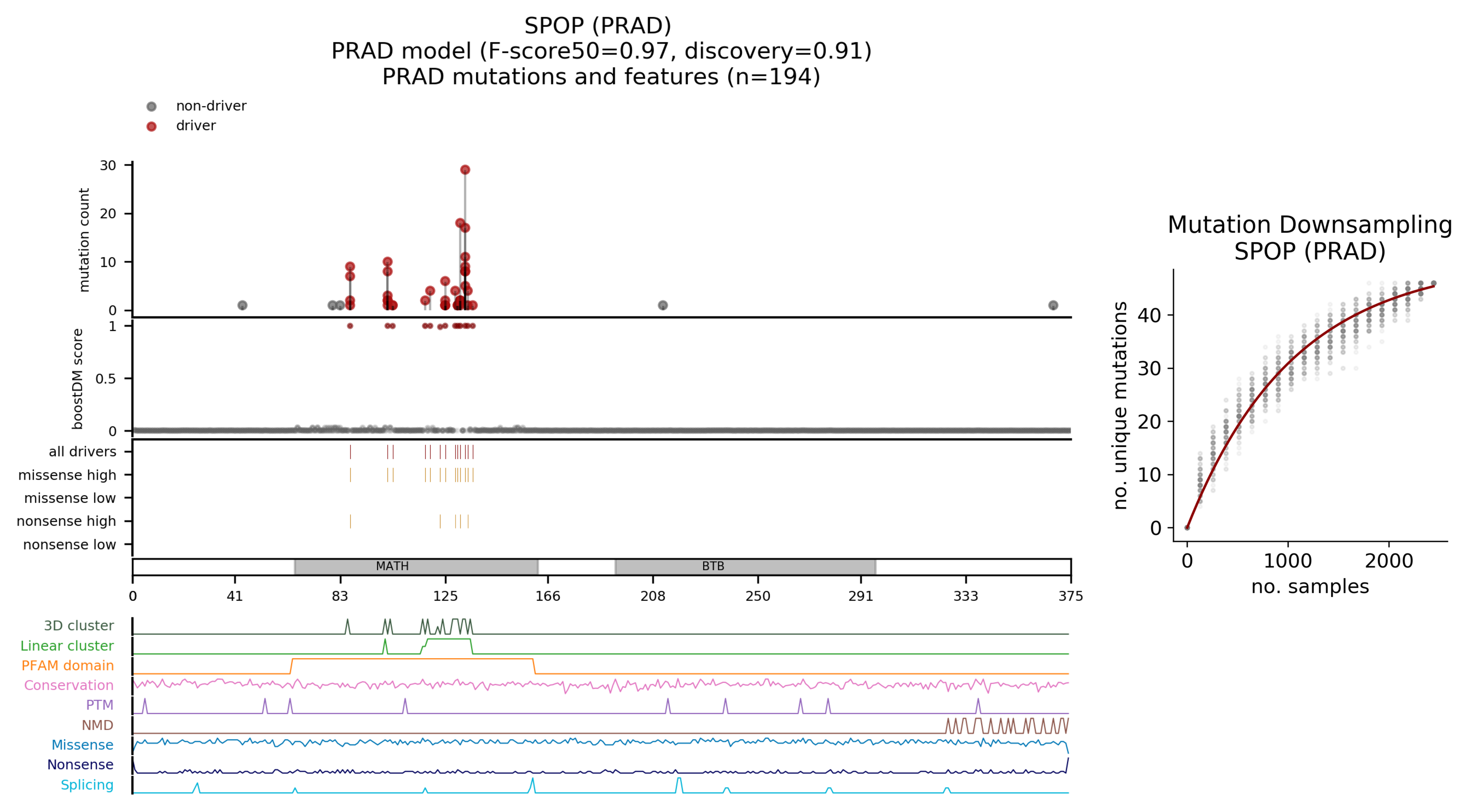
In-silico saturation mutagenesis Left: In silico saturation mutagenesis of all coding variants represented in their relative position of the gene coding sequence. Relevant functional protein domains are represented within each gene body. Potential driver mutations appear in red and potential passenger mutations in gray. The concentration of driver mutations at different regions of the protein is represented as a density. The vertical bar plot shows the frequency bins of boostDM scores. Right: For each random subsample of the tumor type cohort, the number of unique mutations mapping to the gene can be counted. The bend of the best fitting curve to the subsampling data is informative of how close the current pool of mutations is from representing all the possible mutations in this gene-tumor type context..
| Model | Mutations | Driver mutations | Driver mutations (%) |
|---|---|---|---|
| PRAD (Prostate Adenocarcinoma) | 3357 | 97 | 2.89 |
Observed mutations in tumors
The mutations needle plot shows the distribution of the observed
mutations along the protein sequence.
| Mutation (GRCh38) | Protein Position | Samples | Samples (%) | Consequence | Driver | Driver score |
|---|